API
Index
Survey.AbstractSurveyDesignSurvey.BootstrapReplicatesSurvey.JackknifeReplicatesSurvey.ReplicateDesignSurvey.SurveyDesignGLM.glmStatistics.meanStatistics.quantileSurvey.bootweightsSurvey.boxplotSurvey.freedman_diaconisSurvey.histSurvey.jackknifeweightsSurvey.load_dataSurvey.plotSurvey.ratioSurvey.standarderrorSurvey.sturgesSurvey.total
Survey.AbstractSurveyDesign — TypeAbstractSurveyDesignSupertype for every survey design type.
The data passed to a survey constructor is modified. To avoid this pass a copy of the data instead of the original.
Survey.SurveyDesign — TypeSurveyDesign <: AbstractSurveyDesignGeneral survey design encompassing a simple random, stratified, cluster or multi-stage design.
In the case of cluster sample, the clusters are chosen by simple random sampling. All individuals in one cluster are sampled. The clusters are considered disjoint and nested.
strata and clusters must be given as columns in data.
Arguments:
data::AbstractDataFrame: the survey dataset (!this gets modified by the constructor).strata::Union{Nothing, Symbol}=nothing: the stratification variable.clusters::Union{Nothing, Symbol, Vector{Symbol}}=nothing: the clustering variable.weights::Union{Nothing, Symbol}=nothing: the sampling weights.popsize::Union{Nothing, Symbol}=nothing: the (expected) survey population size.
julia> apiclus1 = load_data("apiclus1");
julia> dclus1 = SurveyDesign(apiclus1; clusters=:dnum, weights=:pw)
SurveyDesign:
data: 183×44 DataFrame
strata: none
cluster: dnum
[637, 637, 637 … 448]
popsize: [507.7049, 507.7049, 507.7049 … 507.7049]
sampsize: [15, 15, 15 … 15]
weights: [33.847, 33.847, 33.847 … 33.847]
allprobs: [0.0295, 0.0295, 0.0295 … 0.0295]Survey.ReplicateDesign — TypeReplicateDesign <: AbstractSurveyDesignSurvey design obtained by replicating an original design using an inference method like bootweights or jackknifeweights. If replicate weights are available, then they can be used to directly create a ReplicateDesign object.
Constructors
ReplicateDesign{ReplicateType}(
data::AbstractDataFrame,
replicate_weights::Vector{Symbol};
clusters::Union{Nothing,Symbol,Vector{Symbol}} = nothing,
strata::Union{Nothing,Symbol} = nothing,
popsize::Union{Nothing,Symbol} = nothing,
weights::Union{Nothing,Symbol} = nothing
) where {ReplicateType <: InferenceMethod}
ReplicateDesign{ReplicateType}(
data::AbstractDataFrame,
replicate_weights::UnitIndex{Int};
clusters::Union{Nothing,Symbol,Vector{Symbol}} = nothing,
strata::Union{Nothing,Symbol} = nothing,
popsize::Union{Nothing,Symbol} = nothing,
weights::Union{Nothing,Symbol} = nothing
) where {ReplicateType <: InferenceMethod}
ReplicateDesign{ReplicateType}(
data::AbstractDataFrame,
replicate_weights::Regex;
clusters::Union{Nothing,Symbol,Vector{Symbol}} = nothing,
strata::Union{Nothing,Symbol} = nothing,
popsize::Union{Nothing,Symbol} = nothing,
weights::Union{Nothing,Symbol} = nothing
) where {ReplicateType <: InferenceMethod}Arguments
ReplicateType must be one of the supported inference types; currently the package supports BootstrapReplicates and JackknifeReplicates. The constructor has the same arguments as SurveyDesign. The only additional argument is replicate_weights, which can be of one of the following types.
Vector{Symbol}: In this case, eachSymbolin the vector should represent a column ofdatacontaining the replicate weights.UnitIndex{Int}: For instance, this could be UnitRange(5:10). This will mean that the replicate weights are contained in columns 5 through 10.Regex: In this case, all the columns ofdatawhich match thisRegexwill be treated as the columns containing the replicate weights.
All the columns containing the replicate weights will be renamed to the form replicate_i, where i ranges from 1 to the number of columns containing the replicate weights.
Examples
Here is an example where the bootweights function is used to create a ReplicateDesign{BootstrapReplicates}.
julia> apistrat = load_data("apistrat");
julia> dstrat = SurveyDesign(apistrat; strata=:stype, weights=:pw);
julia> bootstrat = bootweights(dstrat; replicates=1000) # creating a ReplicateDesign using bootweights
ReplicateDesign{BootstrapReplicates}:
data: 200×1044 DataFrame
strata: stype
[E, E, E … H]
cluster: none
popsize: [4420.9999, 4420.9999, 4420.9999 … 755.0]
sampsize: [100, 100, 100 … 50]
weights: [44.21, 44.21, 44.21 … 15.1]
allprobs: [0.0226, 0.0226, 0.0226 … 0.0662]
type: bootstrap
replicates: 1000
If the replicate weights are given to us already, then we can directly pass them to the ReplicateDesign constructor. For instance, in the above example, suppose we had the bootstrat data as a CSV file (for this example, we also rename the columns containing the replicate weights to the form r_i).
julia> using CSV;
julia> DataFrames.rename!(bootstrat.data, ["replicate_"*string(index) => "r_"*string(index) for index in 1:1000]);
julia> CSV.write("apistrat_withreplicates.csv", bootstrat.data);
We can now pass the replicate weights directly to the ReplicateDesign constructor, either as a Vector{Symbol}, a UnitRange or a Regex.
julia> bootstrat_direct = ReplicateDesign{BootstrapReplicates}(CSV.read("apistrat_withreplicates.csv", DataFrame), [Symbol("r_"*string(replicate)) for replicate in 1:1000]; strata=:stype, weights=:pw)
ReplicateDesign{BootstrapReplicates}:
data: 200×1044 DataFrame
strata: stype
[E, E, E … H]
cluster: none
popsize: [4420.9999, 4420.9999, 4420.9999 … 755.0]
sampsize: [100, 100, 100 … 50]
weights: [44.21, 44.21, 44.21 … 15.1]
allprobs: [0.0226, 0.0226, 0.0226 … 0.0662]
type: bootstrap
replicates: 1000
julia> bootstrat_unitrange = ReplicateDesign{BootstrapReplicates}(CSV.read("apistrat_withreplicates.csv", DataFrame), UnitRange(45:1044);strata=:stype, weights=:pw)
ReplicateDesign{BootstrapReplicates}:
data: 200×1044 DataFrame
strata: stype
[E, E, E … H]
cluster: none
popsize: [4420.9999, 4420.9999, 4420.9999 … 755.0]
sampsize: [100, 100, 100 … 50]
weights: [44.21, 44.21, 44.21 … 15.1]
allprobs: [0.0226, 0.0226, 0.0226 … 0.0662]
type: bootstrap
replicates: 1000
julia> bootstrat_regex = ReplicateDesign{BootstrapReplicates}(CSV.read("apistrat_withreplicates.csv", DataFrame), r"r_\d";strata=:stype, weights=:pw)
ReplicateDesign{BootstrapReplicates}:
data: 200×1044 DataFrame
strata: stype
[E, E, E … H]
cluster: none
popsize: [4420.9999, 4420.9999, 4420.9999 … 755.0]
sampsize: [100, 100, 100 … 50]
weights: [44.21, 44.21, 44.21 … 15.1]
allprobs: [0.0226, 0.0226, 0.0226 … 0.0662]
type: bootstrap
replicates: 1000
Survey.BootstrapReplicates — TypeBootstrapReplicates <: InferenceMethodType for the bootstrap replicates method. For more details, see bootweights.
Survey.JackknifeReplicates — TypeJackknifeReplicates <: InferenceMethodType for the Jackknife replicates method. For more details, see jackknifeweights.
Survey.load_data — Functionload_data(name)Load a sample dataset as a DataFrame.
All available datasets can be found here.
julia> apisrs = load_data("apisrs")
200×40 DataFrame
Row │ Column1 cds stype name sname ⋯
│ Int64 Int64 String1 String15 String ⋯
─────┼──────────────────────────────────────────────────────────────────────────
1 │ 1039 15739081534155 H McFarland High McFarland High ⋯
2 │ 1124 19642126066716 E Stowers (Cecil Stowers (Cecil B.) E
3 │ 2868 30664493030640 H Brea-Olinda Hig Brea-Olinda High
4 │ 1273 19644516012744 E Alameda Element Alameda Elementary
5 │ 4926 40688096043293 E Sunnyside Eleme Sunnyside Elementary ⋯
6 │ 2463 19734456014278 E Los Molinos Ele Los Molinos Elementa
7 │ 2031 19647336058200 M Northridge Midd Northridge Middle
8 │ 1736 19647336017271 E Glassell Park E Glassell Park Elemen
⋮ │ ⋮ ⋮ ⋮ ⋮ ⋮ ⋱
194 │ 4880 39686766042782 E Tyler Skills El Tyler Skills Element ⋯
195 │ 993 15636851531987 H Desert Junior/S Desert Junior/Senior
196 │ 969 15635291534775 H North High North High
197 │ 1752 19647336017446 E Hammel Street E Hammel Street Elemen
198 │ 4480 37683386039143 E Audubon Element Audubon Elementary ⋯
199 │ 4062 36678196036222 E Edison Elementa Edison Elementary
200 │ 2683 24657716025621 E Franklin Elemen Franklin Elementary
36 columns and 185 rows omittedSurvey.bootweights — FunctionUse bootweights to create replicate weights using Rao-Wu bootstrap. The function accepts a SurveyDesign and returns a ReplicateDesign{BootstrapReplicates} which has additional columns for replicate weights.
The replicate weight for replicate $r$ is computed using the formula $w_{i}(r) = w_i \times \frac{n_h}{n_h - 1} m_{hj}(r)$ for observation $i$ in psu $j$ of stratum $h$.
In the formula above, $w_i$ is the original weight for observation $i$, $n_h$ is the number of psus in stratum $h$, and $m_{hj}(r)$ is the number of psus in stratum $h$ that are selected in replicate $r$.
julia> using Random
julia> apiclus1 = load_data("apiclus1");
julia> dclus1 = SurveyDesign(apiclus1; clusters = :dnum, popsize=:fpc);
julia> bootweights(dclus1; replicates=1000, rng=MersenneTwister(111)) # choose a seed for deterministic results
ReplicateDesign{BootstrapReplicates}:
data: 183×1044 DataFrame
strata: none
cluster: dnum
[61, 61, 61 … 815]
popsize: [757, 757, 757 … 757]
sampsize: [15, 15, 15 … 15]
weights: [50.4667, 50.4667, 50.4667 … 50.4667]
allprobs: [0.0198, 0.0198, 0.0198 … 0.0198]
type: bootstrap
replicates: 1000
Reference
pg 385, Section 9.3.3 Bootstrap - Sharon Lohr, Sampling Design and Analysis (2010)
Survey.jackknifeweights — Functionjackknifeweights(design::SurveyDesign)Delete-1 Jackknife algorithm for replication weights from sampling weights. The replicate weights are calculated using the following formula.
\[w_{i(hj)} = \begin{cases} w_i\quad\quad &\text{if observation unit }i\text{ is not in stratum }h\\ 0\quad\quad &\text{if observation unit }i\text{ is in psu }j\text{of stratum }h\\ \dfrac{n_h}{n_h - 1}w_i \quad\quad &\text{if observation unit }i\text{ is in stratum }h\text{ but not in psu }j\\ \end{cases}\]
In the above formula, $w_i$ represent the original weights, $w_{i(hj)}$ represent the replicate weights when the $j$th PSU from cluster $h$ is removed, and $n_h$ represents the number of unique PSUs within cluster $h$. Replicate weights are added as columns to design.data, and these columns have names of the form replicate_i, where i ranges from 1 to the number of replicate weight columns.
Examples
julia> using Survey;
julia> apistrat = load_data("apistrat");
julia> dstrat = SurveyDesign(apistrat; strata=:stype, weights=:pw);
julia> rstrat = jackknifeweights(dstrat)
ReplicateDesign{JackknifeReplicates}:
data: 200×244 DataFrame
strata: stype
[E, E, E … M]
cluster: none
popsize: [4420.9999, 4420.9999, 4420.9999 … 1018.0]
sampsize: [100, 100, 100 … 50]
weights: [44.21, 44.21, 44.21 … 20.36]
allprobs: [0.0226, 0.0226, 0.0226 … 0.0491]
type: jackknife
replicates: 200
Reference
pg 380-382, Section 9.3.2 Jackknife - Sharon Lohr, Sampling Design and Analysis (2010)
Survey.standarderror — Functionstandarderror(x::Union{Symbol, Vector{Symbol}}, func::Function, design::ReplicateDesign{BootstrapReplicates}, args...; kwargs...)
Compute the standard error of the estimated mean using the bootstrap method.
Arguments
x::Union{Symbol, Vector{Symbol}}: Symbol or vector of symbols representing the variable(s) for which the mean is estimated.func::Function: Function used to calculate the mean.design::ReplicateDesign{BootstrapReplicates}: Replicate design object.args...: Additional arguments to be passed to the function.kwargs...: Additional keyword arguments.
Returns
df: DataFrame containing the estimated mean and its standard error.
The standard error is calculated using the formula
\[\hat{V}(\hat{\theta}) = \dfrac{1}{R}\sum_{i = 1}^R(\theta_i - \hat{\theta})^2\]
where above $R$ is the number of replicate weights, $\theta_i$ is the estimator computed using the $i$th set of replicate weights, and $\hat{\theta}$ is the estimator computed using the original weights.
Examples
julia> my_mean(df::DataFrame, column, weights) = StatsBase.mean(df[!, column], StatsBase.weights(df[!, weights]));
julia> Survey.standarderror(:api00, my_mean, bclus1)
1×2 DataFrame
Row │ estimator SE
│ Float64 Float64
─────┼────────────────────
1 │ 644.169 23.4107standarderror(x::Symbol, func::Function, design::ReplicateDesign{JackknifeReplicates})
Compute standard error of column x for the given func using the Jackknife method. The formula to compute this variance is the following.
\[\hat{V}_{\text{JK}}(\hat{\theta}) = \sum_{h = 1}^H \dfrac{n_h - 1}{n_h}\sum_{j = 1}^{n_h}(\hat{\theta}_{(hj)} - \hat{\theta})^2\]
Above, $\hat{\theta}$ represents the estimator computed using the original weights, and $\hat{\theta_{(hj)}}$ represents the estimator computed from the replicate weights obtained when PSU $j$ from cluster $h$ is removed.
Examples
julia> my_mean(df::DataFrame, column, weights) = StatsBase.mean(df[!, column], StatsBase.weights(df[!, weights]));
julia> Survey.standarderror(:api00, my_mean, rstrat)
1×2 DataFrame
Row │ estimator SE
│ Float64 Float64
─────┼────────────────────
1 │ 662.287 9.53613Reference
pg 380-382, Section 9.3.2 Jackknife - Sharon Lohr, Sampling Design and Analysis (2010)
Statistics.mean — Functionmean(var, design)Estimate the mean of a variable.
julia> apiclus1 = load_data("apiclus1");
julia> dclus1 = SurveyDesign(apiclus1; clusters = :dnum, weights = :pw)
SurveyDesign:
data: 183×44 DataFrame
strata: none
cluster: dnum
[637, 637, 637 … 448]
popsize: [507.7049, 507.7049, 507.7049 … 507.7049]
sampsize: [15, 15, 15 … 15]
weights: [33.847, 33.847, 33.847 … 33.847]
allprobs: [0.0295, 0.0295, 0.0295 … 0.0295]
julia> mean(:api00, dclus1)
1×1 DataFrame
Row │ mean
│ Float64
─────┼─────────
1 │ 644.169 mean(x::Symbol, design::ReplicateDesign)Compute the standard error of the estimated mean using replicate weights.
Arguments
x::Symbol: Symbol representing the variable for which the mean is estimated.design::ReplicateDesign: Replicate design object.
Returns
df: DataFrame containing the estimated mean and its standard error.
Examples
julia> mean(:api00, bclus1)
1×2 DataFrame
Row │ mean SE
│ Float64 Float64
─────┼──────────────────
1 │ 644.169 23.4107Estimate the mean of a list of variables.
julia> mean([:api00, :enroll], dclus1)
2×2 DataFrame
Row │ names mean
│ String Float64
─────┼─────────────────
1 │ api00 644.169
2 │ enroll 549.716Use replicate weights to compute the standard error of the estimated means.
julia> mean([:api00, :enroll], bclus1)
2×3 DataFrame
Row │ names mean SE
│ String Float64 Float64
─────┼──────────────────────────
1 │ api00 644.169 23.4107
2 │ enroll 549.716 45.7835 mean(var, domain, design)Estimate means of domains.
julia> mean(:api00, :cname, dclus1)
11×2 DataFrame
Row │ mean cname
│ Float64 String
─────┼──────────────────────
1 │ 669.0 Alameda
2 │ 472.0 Fresno
3 │ 452.5 Kern
4 │ 647.267 Los Angeles
5 │ 623.25 Mendocino
6 │ 519.25 Merced
7 │ 710.563 Orange
8 │ 709.556 Plumas
9 │ 659.436 San Diego
10 │ 551.189 San Joaquin
11 │ 732.077 Santa ClaraUse the replicate design to compute standard errors of the estimated means.
julia> mean(:api00, :cname, bclus1)
11×3 DataFrame
Row │ mean SE cname
│ Float64 Float64 String
─────┼────────────────────────────────────
1 │ 732.077 58.2169 Santa Clara
2 │ 659.436 2.66703 San Diego
3 │ 519.25 2.28936e-15 Merced
4 │ 647.267 47.6233 Los Angeles
5 │ 710.563 2.19826e-13 Orange
6 │ 472.0 1.13687e-13 Fresno
7 │ 709.556 1.26058e-13 Plumas
8 │ 669.0 1.27527e-13 Alameda
9 │ 551.189 2.18162e-13 San Joaquin
10 │ 452.5 0.0 Kern
11 │ 623.25 1.09545e-13 MendocinoSurvey.total — Functiontotal(var, design)Estimate the population total of variable.
julia> apiclus1 = load_data("apiclus1");
julia> dclus1 = SurveyDesign(apiclus1; clusters = :dnum, weights = :pw);
julia> total(:api00, dclus1)
1×1 DataFrame
Row │ total
│ Float64
─────┼───────────
1 │ 3.98999e6total(x::Symbol, design::ReplicateDesign)Compute the standard error of the estimated total using replicate weights.
Arguments
x::Symbol: Symbol representing the variable for which the total is estimated.design::ReplicateDesign: Replicate design object.
Returns
df: DataFrame containing the estimated total and its standard error.
Examples
julia> total(:api00, bclus1)
1×2 DataFrame
Row │ total SE
│ Float64 Float64
─────┼──────────────────────
1 │ 3.98999e6 9.01611e5Estimate the population total of a list of variables.
julia> total([:api00, :enroll], dclus1)
2×2 DataFrame
Row │ names total
│ String Float64
─────┼───────────────────
1 │ api00 3.98999e6
2 │ enroll 3.40494e6Use replicate weights to compute the standard error of the estimated means.
julia> total([:api00, :enroll], bclus1)
2×3 DataFrame
Row │ names total SE
│ String Float64 Float64
─────┼──────────────────────────────
1 │ api00 3.98999e6 9.01611e5
2 │ enroll 3.40494e6 9.33396e5 total(var, domain, design)Estimate population totals of domains.
julia> total(:api00, :cname, dclus1)
11×2 DataFrame
Row │ total cname
│ Float64 String
─────┼─────────────────────────────
1 │ 249080.0 Alameda
2 │ 63903.1 Fresno
3 │ 30631.5 Kern
4 │ 3.2862e5 Los Angeles
5 │ 84380.6 Mendocino
6 │ 70300.2 Merced
7 │ 3.84807e5 Orange
8 │ 2.16147e5 Plumas
9 │ 1.2276e6 San Diego
10 │ 6.90276e5 San Joaquin
11 │ 6.44244e5 Santa ClaraUse the replicate design to compute standard errors of the estimated totals.
julia> total(:api00, :cname, bclus1)
11×3 DataFrame
Row │ total SE cname
│ Float64 Float64 String
─────┼────────────────────────────────────────────
1 │ 6.44244e5 4.2273e5 Santa Clara
2 │ 1.2276e6 8.62727e5 San Diego
3 │ 70300.2 71336.3 Merced
4 │ 3.2862e5 2.93936e5 Los Angeles
5 │ 3.84807e5 3.88014e5 Orange
6 │ 63903.1 64781.7 Fresno
7 │ 2.16147e5 2.12089e5 Plumas
8 │ 249080.0 2.49228e5 Alameda
9 │ 6.90276e5 6.81604e5 San Joaquin
10 │ 30631.5 30870.3 Kern
11 │ 84380.6 80215.9 MendocinoStatistics.quantile — Functionquantile(var, design, p; kwargs...)Estimate quantile of a variable.
Hyndman and Fan compiled a taxonomy of nine algorithms to estimate quantiles. These are implemented in Statistics.quantile, which this function calls. Julia, R and Python-numpy use the same defaults
References:
- Hyndman, R.J and Fan, Y. (1996) "Sample Quantiles in Statistical Packages", The American Statistician, Vol. 50, No. 4, pp. 361-365.
- Quantiles on wikipedia
- Complex Surveys: a guide to analysis using R, Section 2.4.1 and Appendix C.4.
julia> apisrs = load_data("apisrs");
julia> srs = SurveyDesign(apisrs; weights=:pw);
julia> quantile(:api00, srs, 0.5)
1×1 DataFrame
Row │ 0.5th percentile
│ Float64
─────┼──────────────────
1 │ 659.0quantile(x::Symbol, design::ReplicateDesign, p; kwargs...)Compute the standard error of the estimated quantile using replicate weights.
Arguments
x::Symbol: Symbol representing the variable for which the quantile is estimated.design::ReplicateDesign: Replicate design object.p::Real: Quantile value to estimate, ranging from 0 to 1.kwargs...: Additional keyword arguments.
Returns
df: DataFrame containing the estimated quantile and its standard error.
Examples
julia> quantile(:api00, bsrs, 0.5)
1×2 DataFrame
Row │ 0.5th percentile SE
│ Float64 Float64
─────┼───────────────────────────
1 │ 659.0 14.9764quantile(var, design, p; kwargs...)Estimate quantiles of a list of variables.
julia> quantile(:enroll, srs, [0.1,0.2,0.5,0.75,0.95])
5×2 DataFrame
Row │ percentile statistic
│ String Float64
─────┼───────────────────────
1 │ 0.1 245.5
2 │ 0.2 317.6
3 │ 0.5 453.0
4 │ 0.75 668.5
5 │ 0.95 1473.1Use replicate weights to compute the standard errors of the estimated quantiles.
julia> quantile(:enroll, bsrs, [0.1,0.2,0.5,0.75,0.95])
5×3 DataFrame
Row │ percentile statistic SE
│ String Float64 Float64
─────┼─────────────────────────────────
1 │ 0.1 245.5 20.2964
2 │ 0.2 317.6 13.5435
3 │ 0.5 453.0 24.9719
4 │ 0.75 668.5 34.2487
5 │ 0.95 1473.1 142.568quantile(var, domain, design)
Estimate a quantile of domains.
julia> quantile(:api00, :cname, dclus1, 0.5)
11×2 DataFrame
Row │ 0.5th percentile cname
│ Float64 String
─────┼───────────────────────────────
1 │ 669.0 Alameda
2 │ 474.5 Fresno
3 │ 452.5 Kern
4 │ 628.0 Los Angeles
5 │ 616.5 Mendocino
6 │ 519.5 Merced
7 │ 717.5 Orange
8 │ 699.0 Plumas
9 │ 657.0 San Diego
10 │ 542.0 San Joaquin
11 │ 718.0 Santa ClaraSurvey.ratio — Functionratio(numerator, denominator, design)Estimate the ratio of the columns specified in numerator and denominator.
julia> apiclus1 = load_data("apiclus1");
julia> dclus1 = SurveyDesign(apiclus1; clusters = :dnum, weights = :pw);
julia> ratio([:api00, :enroll], dclus1)
1×1 DataFrame
Row │ ratio
│ Float64
─────┼─────────
1 │ 1.17182
ratio(x::Vector{Symbol}, design::ReplicateDesign)Compute the standard error of the ratio using replicate weights.
Arguments
variable_num::Symbol: Symbol representing the numerator variable.variable_den::Symbol: Symbol representing the denominator variable.design::ReplicateDesign: Replicate design object.
Examples
julia> ratio([:api00, :api99], bclus1)
1×2 DataFrame
Row │ estimator SE
│ Float64 Float64
─────┼───────────────────────
1 │ 1.06127 0.00672259ratio(var, domain, design)Estimate ratios of domains.
julia> ratio([:api00, :api99], :cname, dclus1)
11×2 DataFrame
Row │ ratio cname
│ Float64 String
─────┼──────────────────────
1 │ 1.09852 Alameda
2 │ 1.17779 Fresno
3 │ 1.11453 Kern
4 │ 1.06307 Los Angeles
5 │ 1.00565 Mendocino
6 │ 1.08121 Merced
7 │ 1.03628 Orange
8 │ 1.02127 Plumas
9 │ 1.06112 San Diego
10 │ 1.07331 San Joaquin
11 │ 1.05598 Santa ClaraUse the replicate design to compute standard errors of the estimated means.
julia> ratio([:api00, :api99], :cname, bclus1)
11×3 DataFrame
Row │ estimator SE cname
│ Float64 Float64 String
─────┼─────────────────────────────────────
1 │ 1.05598 0.0189429 Santa Clara
2 │ 1.06112 0.00979481 San Diego
3 │ 1.08121 6.85453e-17 Merced
4 │ 1.06307 0.0257137 Los Angeles
5 │ 1.03628 0.0 Orange
6 │ 1.17779 6.27535e-18 Fresno
7 │ 1.02127 0.0 Plumas
8 │ 1.09852 2.12683e-16 Alameda
9 │ 1.07331 2.22045e-16 San Joaquin
10 │ 1.11453 0.0 Kern
11 │ 1.00565 0.0 MendocinoGLM.glm — Functionglm(formula::FormulaTerm, design::ReplicateDesign, args...; kwargs...)Perform generalized linear modeling (GLM) using the survey design with replicates.
Arguments
formula: AFormulaTermspecifying the model formula.design: AReplicateDesignobject representing the survey design with replicates.args...: Additional arguments to be passed to theglmfunction.kwargs...: Additional keyword arguments to be passed to theglmfunction.
Returns
A DataFrame containing the estimates for model coefficients and their standard errors.
Example
apisrs = load_data("apisrs")
srs = SurveyDesign(apisrs)
bsrs = bootweights(srs, replicates = 2000)
result = glm(@formula(api00 ~ api99), bsrs, Normal())julia> glm(@formula(api00 ~ api99), bsrs, Normal())
2×2 DataFrame
Row │ estimator SE
│ Float64 Float64
─────┼──────────────────────
1 │ 63.2831 9.41231
2 │ 0.949762 0.0135488Survey.plot — Functionplot(design, x, y; kwargs...)Scatter plot of survey design variables x and y.
The plot takes into account the frequency weights specified by the user in the design.
julia> using AlgebraOfGraphics
julia> apisrs = load_data("apisrs");
julia> srs = SurveyDesign(apisrs; weights=:pw);
julia> s = plot(srs, :api99, :api00);
julia> save("scatter.png", s)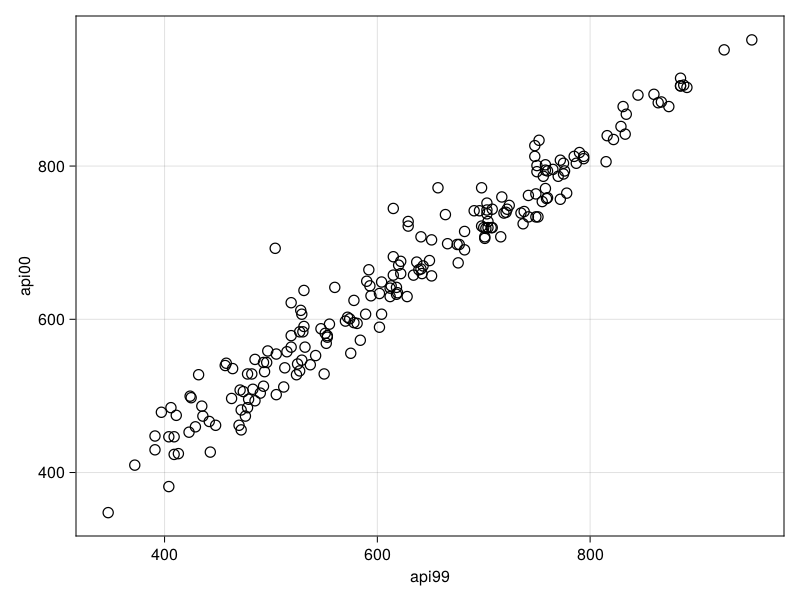
Survey.boxplot — Functionboxplot(design, x, y; kwargs...)Box plot of survey design variable y grouped by column x.
Weights can be specified by a Symbol using the keyword argument weights.
The keyword arguments are all the arguments that can be passed to mapping in AlgebraOfGraphics.
julia> using AlgebraOfGraphics
julia> apisrs = load_data("apisrs");
julia> srs = SurveyDesign(apisrs; weights=:pw);
julia> bp = boxplot(srs, :stype, :enroll; weights = :pw);
julia> save("boxplot.png", bp)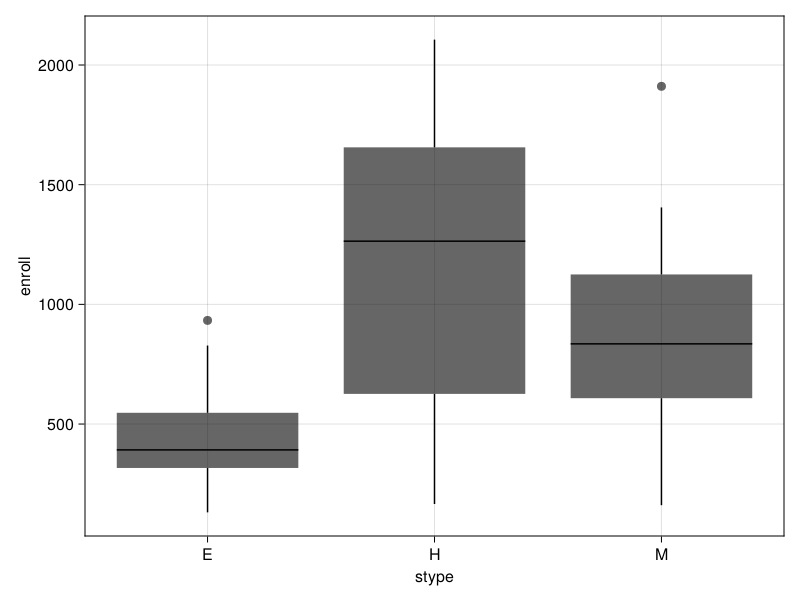
Survey.hist — Functionhist(design, var, bins = freedman_diaconis; normalization = :density, kwargs...)Histogram plot of a survey design variable given by var.
bins can be an Integer specifying the number of equal-width bins, an AbstractVector specifying the bins intervals, or a Function specifying the function used for calculating the number of bins. The possible functions are sturges and freedman_diaconis.
The normalization can be set to :none, :density, :probability or :pdf. See AlgebraOfGraphics.histogram for more information.
For the complete argument list see Makie.hist.
The weights argument should be a Symbol specifying a design variable.
julia> using AlgebraOfGraphics
julia> apisrs = load_data("apisrs");
julia> srs = SurveyDesign(apisrs; weights=:pw);
julia> h = hist(srs, :enroll);
julia> save("hist.png", h)
Survey.sturges — Functionsturges(design::SurveyDesign, var::Symbol)Calculate the number of bins to use in a histogram using the Sturges rule.
Examples
julia> apisrs = load_data("apisrs");
julia> srs = SurveyDesign(apisrs; weights=:pw);
julia> sturges(srs, :enroll)
9Survey.freedman_diaconis — Functionfreedman_diaconis(design::SurveyDesign, var::Symbol)Calculate the number of bins to use in a histogram using the Freedman-Diaconis rule.
Examples
julia> apisrs = load_data("apisrs");
julia> srs = SurveyDesign(apisrs; weights=:pw);
julia> freedman_diaconis(srs, :enroll)
18